Efficient Information Retrieval RAG for Complex PDFs Using RAPTOR

RAPTOR introduces a novel approach to retrieval-augmented language models by constructing a recursive tree structure from documents. This allows for more efficient and context-aware information retrieval across large texts, addressing common limitations in traditional language models.
For detailed methodologies and implementations, refer to the original paper:
What is RAPTOR?
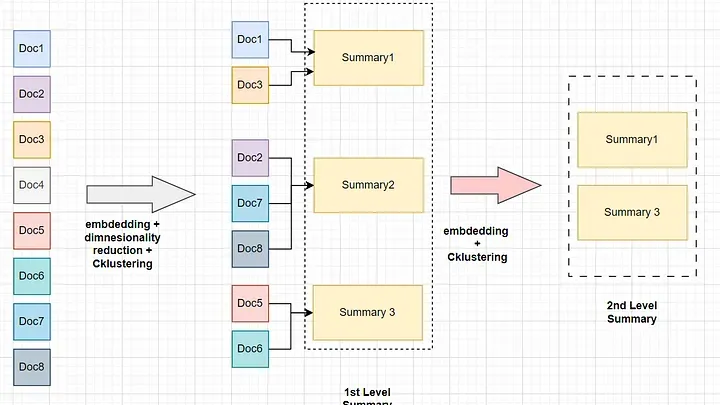
Suppose we have 8 document chunks that belong to one large handbook. Instead of just embedding the chunks and performing retrieval on them, we embed the chunks and then run a dimensionality reduction on them as it would be computationally expensive to generate clusters for all the dimension which is 1536 in case of OpenAI embeddings and 384 in case of common open-source small embedding models.
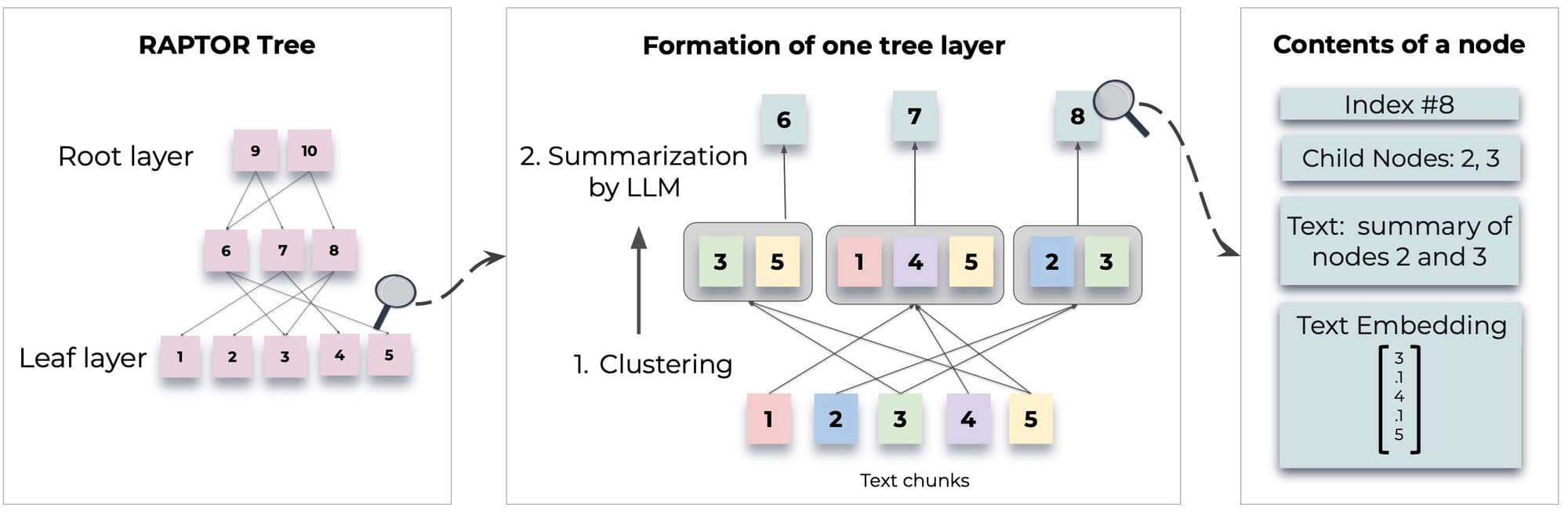
Then cluster the reduced dimension with a clustering algorithm.We then take all the chunks that belong to each cluster and summarize the context for each clusters. The generated summaries are gain embedded and clustered repeating the process until the token limit (context window) of the model is reached.
In short, the intuition behind RAPTOR as follows:
- cluster and summarize similar documents.
- capture information from related documents into a summary.
- provide help on questions that need content from a fewer context to answer.
Choosing Between - Tree Traversal Retrieval vs. Collapsed Tree Retrieval.
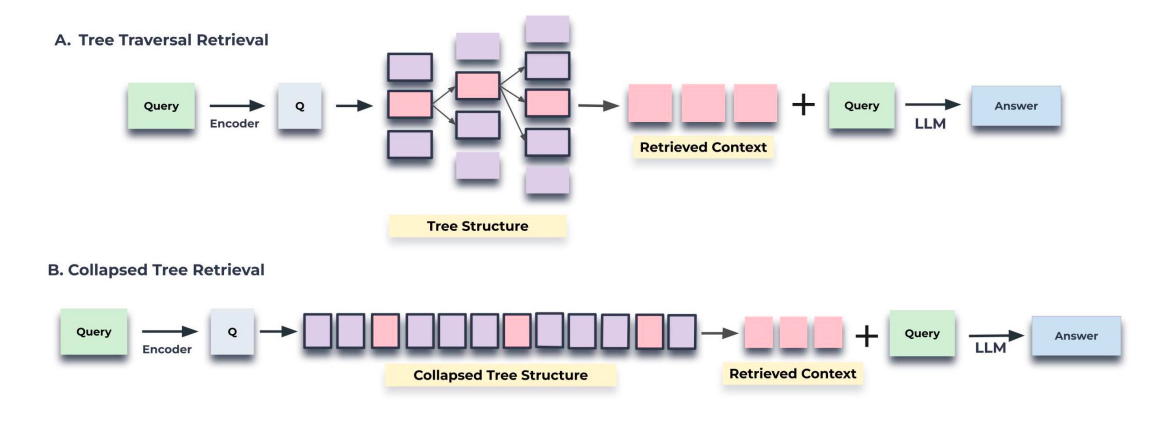
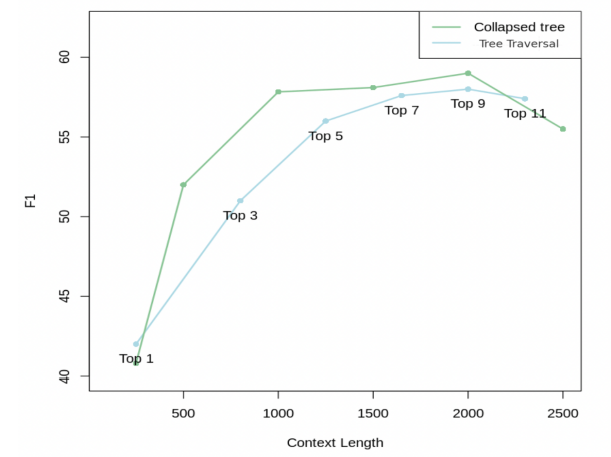
The collapsed tree approach is preferred due to its enhanced flexibility and superior performance compared to traditional tree traversal methods. By collapsing the tree and searching all nodes simultaneously, it allows for dynamic retrieval of information at varying levels of granularity tailored to specific questions. This flexibility ensures that RAPTOR can adaptively select nodes across different layers of the tree, optimizing relevance and comprehensiveness in information retrieval tasks. Despite requiring cosine similarity searches across all nodes, efficiencies can be achieved using fast k-nearest neighbor. Overall, the collapsed tree method with 2000 maximum tokens provides optimal performance by accommodating varying token counts across nodes and aligning with model context constraints.
In this Blog, we have presented RAPTOR, a novel tree-based retrieval system that augments the parametric knowledge of large language models with contextual information at various levels of abstraction. By employing recursive clustering and summarization techniques, RAPTOR creates a hierarchical tree structure that is capable of synthesizing information across various sections of the retrieval corpora. During the query phase, RAPTOR leverages this tree structure for more effective retrieval. Our controlled experiments demonstrated that RAPTOR not only outperforms traditional retrieval methods but also sets new performance benchmarks on several question-answering tasks.
Key Features
Text Extraction
The system can efficiently extract and process text from PDFs, ensuring accurate and comprehensive information retrieval. This feature is particularly useful for extracting large text blocks or specific sections from complex documents.
Table Extraction
RAG-RAPTOR-DEMO excels at identifying and parsing tables within PDFs, allowing for the retrieval of structured data. This capability is crucial for answering data-specific questions and extracting numerical or categorical data efficiently
Image Analysis
RAG-RAPTOR-DEMO also offers the ability to extract and interpret images within PDFs. By providing contextually relevant information, this feature enhances the overall understanding of the document's content.
Technologies Used
The RAG-RAPTOR-DEMO project leverages several advanced technologies:
- LangChain: A framework for building applications with language models.
- RAG (Retrieval-Augmented Generation): Combines retrieval and generation for more accurate answers.
- RAPTOR: Constructs a recursive tree structure for efficient, context-aware information retrieval.
- Streamlit: A framework for creating interactive web applications with Python.
- Unstructured.io: A tool for parsing and extracting complex content from PDFs, such as tables, graphs, and images.
- Poetry: A dependency management and packaging tool for Python.
Code Implementation:
To start using RAPTOR, you begin by generating your document chunk using Unstructured.io. Follow the steps outlined in the blog provided here. Once you've created your chunk, pass it to RAPTOR, which will process it and return the resulting RAPTOR chunk.
Tree Construction
The clustering approach in tree construction includes a few interesting ideas.
GMM (Gaussian Mixture Model)
- Model the distribution of data points across different clusters
- Optimal number of clusters by evaluating the model's Bayesian Information Criterion (BIC)
UMAP (Uniform Manifold Approximation and Projection)
- Supports clustering
- Reduces the dimensionality of high-dimensional data
- UMAP helps to highlight the natural grouping of data points based on their similarities
Local and Global Clustering
- Used to analyze data at different scales
- Both fine-grained and broader patterns within the data are captured effectively
Thresholding
- Apply in the context of GMM to determine cluster membership
- Based on the probability distribution (assignment of data points to ≥ 1 cluster)
The below raptor.py Python script provides a comprehensive framework for embedding, clustering, and summarizing text documents using various machine learning techniques. Here’s a breakdown of its components and functionality:
import umap
import numpy as np
import pandas as pd
from typing import Dict, List, Optional, Tuple
from langchain.prompts import ChatPromptTemplate
from langchain_core.output_parsers import StrOutputParser
from sklearn.mixture import GaussianMixture
from langchain_openai import OpenAIEmbeddings
from langchain_openai import ChatOpenAI
RANDOM_SEED = 224
embd = OpenAIEmbeddings()
model = ChatOpenAI(temperature=0, model="gpt-4o")
def global_cluster_embeddings(
embeddings: np.ndarray,
dim: int,
n_neighbors: Optional[int] = None,
metric: str = "cosine",
) -> np.ndarray:
"""
Perform global dimensionality reduction on the embeddings using UMAP.
Parameters:
- embeddings: The input embeddings as a numpy array.
- dim: The target dimensionality for the reduced space.
- n_neighbors: Optional; the number of neighbors to consider for each point.
If not provided, it defaults to the square root of the number of embeddings.
- metric: The distance metric to use for UMAP.
Returns:
- A numpy array of the embeddings reduced to the specified dimensionality.
"""
if n_neighbors is None:
n_neighbors = int((len(embeddings) - 1) ** 0.5)
return umap.UMAP(
n_neighbors=n_neighbors, n_components=dim, metric=metric
).fit_transform(embeddings)
def local_cluster_embeddings(
embeddings: np.ndarray, dim: int, num_neighbors: int = 10, metric: str = "cosine"
) -> np.ndarray:
"""
Perform local dimensionality reduction on the embeddings using UMAP, typically after global clustering.
Parameters:
- embeddings: The input embeddings as a numpy array.
- dim: The target dimensionality for the reduced space.
- num_neighbors: The number of neighbors to consider for each point.
- metric: The distance metric to use for UMAP.
Returns:
- A numpy array of the embeddings reduced to the specified dimensionality.
"""
return umap.UMAP(
n_neighbors=num_neighbors, n_components=dim, metric=metric
).fit_transform(embeddings)
def get_optimal_clusters(
embeddings: np.ndarray, max_clusters: int = 50, random_state: int = RANDOM_SEED
) -> int:
"""
Determine the optimal number of clusters using the Bayesian Information Criterion (BIC) with a Gaussian Mixture Model.
Parameters:
- embeddings: The input embeddings as a numpy array.
- max_clusters: The maximum number of clusters to consider.
- random_state: Seed for reproducibility.
Returns:
- An integer representing the optimal number of clusters found.
"""
max_clusters = min(max_clusters, len(embeddings))
n_clusters = np.arange(1, max_clusters)
bics = []
for n in n_clusters:
gm = GaussianMixture(n_components=n, random_state=random_state)
gm.fit(embeddings)
bics.append(gm.bic(embeddings))
return n_clusters[np.argmin(bics)]
def GMM_cluster(embeddings: np.ndarray, threshold: float, random_state: int = 0):
"""
Cluster embeddings using a Gaussian Mixture Model (GMM) based on a probability threshold.
Parameters:
- embeddings: The input embeddings as a numpy array.
- threshold: The probability threshold for assigning an embedding to a cluster.
- random_state: Seed for reproducibility.
Returns:
- A tuple containing the cluster labels and the number of clusters determined.
"""
n_clusters = get_optimal_clusters(embeddings)
gm = GaussianMixture(n_components=n_clusters, random_state=random_state)
gm.fit(embeddings)
probs = gm.predict_proba(embeddings)
labels = [np.where(prob > threshold)[0] for prob in probs]
return labels, n_clusters
def perform_clustering(
embeddings: np.ndarray,
dim: int,
threshold: float,
) -> List[np.ndarray]:
"""
Perform clustering on the embeddings by first reducing their dimensionality globally, then clustering
using a Gaussian Mixture Model, and finally performing local clustering within each global cluster.
Parameters:
- embeddings: The input embeddings as a numpy array.
- dim: The target dimensionality for UMAP reduction.
- threshold: The probability threshold for assigning an embedding to a cluster in GMM.
Returns:
- A list of numpy arrays, where each array contains the cluster IDs for each embedding.
"""
if len(embeddings) <= dim + 1:
# Avoid clustering when there's insufficient data
return [np.array([0]) for _ in range(len(embeddings))]
# Global dimensionality reduction
reduced_embeddings_global = global_cluster_embeddings(embeddings, dim)
# Global clustering
global_clusters, n_global_clusters = GMM_cluster(
reduced_embeddings_global, threshold
)
all_local_clusters = [np.array([]) for _ in range(len(embeddings))]
total_clusters = 0
# Iterate through each global cluster to perform local clustering
for i in range(n_global_clusters):
# Extract embeddings belonging to the current global cluster
global_cluster_embeddings_ = embeddings[
np.array([i in gc for gc in global_clusters])
]
if len(global_cluster_embeddings_) == 0:
continue
if len(global_cluster_embeddings_) <= dim + 1:
# Handle small clusters with direct assignment
local_clusters = [np.array([0]) for _ in global_cluster_embeddings_]
n_local_clusters = 1
else:
# Local dimensionality reduction and clustering
reduced_embeddings_local = local_cluster_embeddings(
global_cluster_embeddings_, dim
)
local_clusters, n_local_clusters = GMM_cluster(
reduced_embeddings_local, threshold
)
# Assign local cluster IDs, adjusting for total clusters already processed
for j in range(n_local_clusters):
local_cluster_embeddings_ = global_cluster_embeddings_[
np.array([j in lc for lc in local_clusters])
]
indices = np.where(
(embeddings == local_cluster_embeddings_[:, None]).all(-1)
)[1]
for idx in indices:
all_local_clusters[idx] = np.append(
all_local_clusters[idx], j + total_clusters
)
total_clusters += n_local_clusters
return all_local_clusters
### --- Our code below --- ###
def embed(texts):
"""
Generate embeddings for a list of text documents.
This function assumes the existence of an `embd` object with a method `embed_documents`
that takes a list of texts and returns their embeddings.
Parameters:
- texts: List[str], a list of text documents to be embedded.
Returns:
- numpy.ndarray: An array of embeddings for the given text documents.
"""
text_embeddings = embd.embed_documents(texts)
text_embeddings_np = np.array(text_embeddings)
return text_embeddings_np
def embed_cluster_texts(texts):
"""
Embeds a list of texts and clusters them, returning a DataFrame with texts, their embeddings, and cluster labels.
This function combines embedding generation and clustering into a single step. It assumes the existence
of a previously defined `perform_clustering` function that performs clustering on the embeddings.
Parameters:
- texts: List[str], a list of text documents to be processed.
Returns:
- pandas.DataFrame: A DataFrame containing the original texts, their embeddings, and the assigned cluster labels.
"""
text_embeddings_np = embed(texts) # Generate embeddings
cluster_labels = perform_clustering(
text_embeddings_np, 10, 0.1
) # Perform clustering on the embeddings
df = pd.DataFrame() # Initialize a DataFrame to store the results
df["text"] = texts # Store original texts
df["embd"] = list(text_embeddings_np) # Store embeddings as a list in the DataFrame
df["cluster"] = cluster_labels # Store cluster labels
return df
def fmt_txt(df: pd.DataFrame) -> str:
"""
Formats the text documents in a DataFrame into a single string.
Parameters:
- df: DataFrame containing the 'text' column with text documents to format.
Returns:
- A single string where all text documents are joined by a specific delimiter.
"""
unique_txt = df["text"].tolist()
return "--- --- \n --- --- ".join(unique_txt)
def embed_cluster_summarize_texts(
texts: List[str], level: int
) -> Tuple[pd.DataFrame, pd.DataFrame]:
"""
Embeds, clusters, and summarizes a list of texts. This function first generates embeddings for the texts,
clusters them based on similarity, expands the cluster assignments for easier processing, and then summarizes
the content within each cluster.
Parameters:
- texts: A list of text documents to be processed.
- level: An integer parameter that could define the depth or detail of processing.
Returns:
- Tuple containing two DataFrames:
1. The first DataFrame (`df_clusters`) includes the original texts, their embeddings, and cluster assignments.
2. The second DataFrame (`df_summary`) contains summaries for each cluster, the specified level of detail,
and the cluster identifiers.
"""
# Embed and cluster the texts, resulting in a DataFrame with 'text', 'embd', and 'cluster' columns
df_clusters = embed_cluster_texts(texts)
# Prepare to expand the DataFrame for easier manipulation of clusters
expanded_list = []
# Expand DataFrame entries to document-cluster pairings for straightforward processing
for index, row in df_clusters.iterrows():
for cluster in row["cluster"]:
expanded_list.append(
{"text": row["text"], "embd": row["embd"], "cluster": cluster}
)
# Create a new DataFrame from the expanded list
expanded_df = pd.DataFrame(expanded_list)
# Retrieve unique cluster identifiers for processing
all_clusters = expanded_df["cluster"].unique()
print(f"--Generated {len(all_clusters)} clusters--")
# Summarization
template = """
Give a detailed summary of the provided context : {context}
"""
prompt = ChatPromptTemplate.from_template(template)
chain = prompt | model | StrOutputParser()
# Format text within each cluster for summarization
summaries = []
for i in all_clusters:
df_cluster = expanded_df[expanded_df["cluster"] == i]
formatted_txt = fmt_txt(df_cluster)
summaries.append(chain.invoke({"context": formatted_txt}))
# Create a DataFrame to store summaries with their corresponding cluster and level
df_summary = pd.DataFrame(
{
"summaries": summaries,
"level": [level] * len(summaries),
"cluster": list(all_clusters),
}
)
return df_clusters, df_summary
def recursive_embed_cluster_summarize(
texts: List[str], level: int = 1, n_levels: int = 3
) -> Dict[int, Tuple[pd.DataFrame, pd.DataFrame]]:
"""
Recursively embeds, clusters, and summarizes texts up to a specified level or until
the number of unique clusters becomes 1, storing the results at each level.
Parameters:
- texts: List[str], texts to be processed.
- level: int, current recursion level (starts at 1).
- n_levels: int, maximum depth of recursion.
Returns:
- Dict[int, Tuple[pd.DataFrame, pd.DataFrame]], a dictionary where keys are the recursion
levels and values are tuples containing the clusters DataFrame and summaries DataFrame at that level.
"""
results = {} # Dictionary to store results at each level
# Perform embedding, clustering, and summarization for the current level
df_clusters, df_summary = embed_cluster_summarize_texts(texts, level)
# Store the results of the current level
results[level] = (df_clusters, df_summary)
# Determine if further recursion is possible and meaningful
unique_clusters = df_summary["cluster"].nunique()
if level < n_levels and unique_clusters > 1:
# Use summaries as the input texts for the next level of recursion
new_texts = df_summary["summaries"].tolist()
next_level_results = recursive_embed_cluster_summarize(
new_texts, level + 1, n_levels
)
# Merge the results from the next level into the current results dictionary
results.update(next_level_results)
return resultsHere’s a breakdown of its components and functionality:
Libraries and Initialization
- Libraries: Imports necessary libraries including
umapfor dimensionality reduction,numpyandpandasfor data manipulation, andsklearnfor Gaussian Mixture Models. - Initialization: Initializes
OpenAIEmbeddings(embd) andChatOpenAI(model) objects for embedding text and generating summaries respectively.
Dimensionality Reduction and Clustering Functions
- Global Clustering (
global_cluster_embeddings):- Uses UMAP for global dimensionality reduction of embeddings.
- Local Clustering (
local_cluster_embeddings):- Applies UMAP for local dimensionality reduction after global clustering.
- Optimal Number of Clusters (
get_optimal_clusters):- Determines the optimal number of clusters using Bayesian Information Criterion (BIC) with Gaussian Mixture Models.
- Gaussian Mixture Model Clustering (
GMM_cluster):- Clusters embeddings using GMM based on a probability threshold.
- Perform Clustering (
perform_clustering):- Integrates global dimensionality reduction, GMM clustering, and local clustering within global clusters.
Text Embedding and Clustering Functions
- Embedding (
embed): Generates embeddings for a list of text documents usingembd. - Embed and Cluster Texts (
embed_cluster_texts): Embeds texts and clusters them based on similarity, returning a DataFrame with text, embeddings, and cluster labels. - Text Formatting (
fmt_txt): Formats text documents into a single string for summarization. - Embed, Cluster, and Summarize Texts (
embed_cluster_summarize_texts): Embeds, clusters, and summarizes texts, generating clusters and their corresponding summaries.
Recursive Summarization Function
- Recursive Embed, Cluster, and Summarize (
recursive_embed_cluster_summarize):- Recursively embeds, clusters, and summarizes texts up to a specified level or until the number of unique clusters becomes 1, storing results at each level.
Summary Generation
- Summarization Template: Utilizes a template-based approach (
ChatPromptTemplate) to generate detailed summaries for clustered texts using GPT-4o.
The below main() function orchestrates a series of steps to process PDF documents, extract text and image data, apply advanced text analysis (like RAPTOR), and finally store the processed data into a PostgreSQL database. Here’s a brief overview and the flow of execution:
from unstructured_ingest import *
def main():
collection_name="a1"
print("started file reader...")
raw_pdf_elements=file_reader()
print("text_insert started...")
text_insert(raw_pdf_elements)
print("image_insert started...")
last_indices=get_last_index_of_page(raw_pdf_elements)
image_insert_with_text(raw_pdf_elements,last_indices)
get_docummets()
print("Raptor started...")
raptor_texts = raptor()
get_documents_with_raptor(raptor_texts)
print("add data to postgres Started...")
add_docs_to_postgres(collection_name)
print("All Done...")
if __name__=="__main__":
main()Steps in main() Function:
- Importing Functions:
- Imports necessary functions from
unstructured_ingest.
- Imports necessary functions from
- Setting Up:
- Defines
collection_namefor PostgreSQL. - Prints status messages for clarity.
- Defines
- File Reading (
file_reader()):- Reads PDF file (
fy2024.pdf) and extracts raw elements (raw_pdf_elements).
- Reads PDF file (
- Text Extraction (
text_insert()):- Processes
raw_pdf_elementsto extract and summarize text content. - Populates
text_elementsandtext_summaries.
- Processes
- Image Extraction with Text (
image_insert_with_text()):- Retrieves last indices of pages from
raw_pdf_elements. - Extracts images from PDF and summarizes associated text using image content.
- Populates
image_elementsandimage_and_text_summaries.
- Retrieves last indices of pages from
- Document Preparation (
get_documents_with_raptor()):- Uses RAPTOR to analyze and prepare document content (
raptor_texts). - Creates
documentswith enriched metadata and content.
- Uses RAPTOR to analyze and prepare document content (
- Storing Data (
add_docs_to_postgres()):- Adds prepared documents to a PostgreSQL database (
collection_name).
- Adds prepared documents to a PostgreSQL database (
Key Components:
- PDF Processing: Utilizes
file_reader()for initial PDF parsing and element extraction. - Text and Image Processing: Uses
text_insert()andimage_insert_with_text()to handle text and image extraction and summarization. - Advanced Analysis: Applies RAPTOR analysis via
raptor()to enhance document content understanding. - Database Integration: Stores processed documents into PostgreSQL using
add_docs_to_postgres().
Conclusion:
This script provides a structured approach to ingest unstructured PDF data, extract meaningful content through text and image analysis, apply advanced text analysis techniques like RAPTOR, and persist processed data into a PostgreSQL database for further analysis or retrieval. Adjustments or extensions to this workflow can be made based on specific project requirements or additional functionalities needed.
Final Result:-
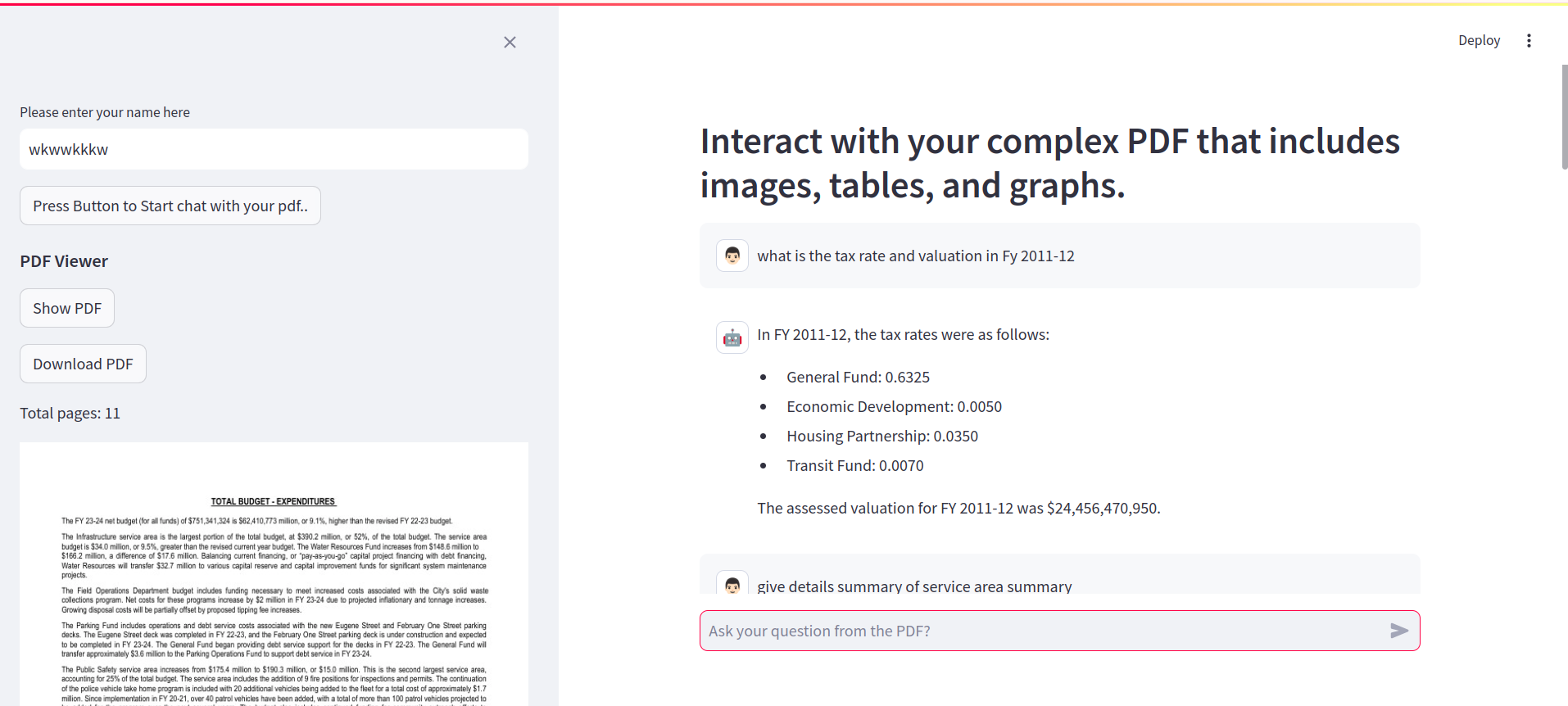
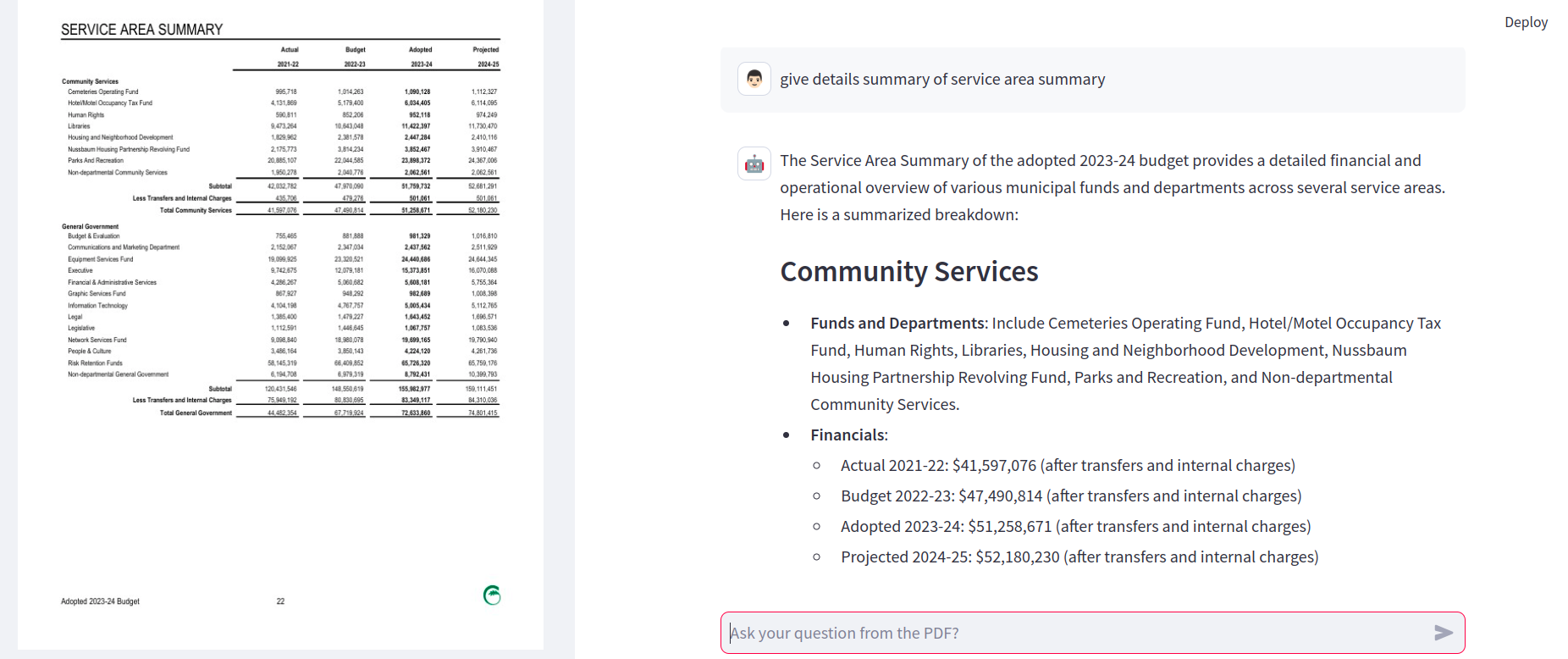
FAQ's
1. What is raptor in rag?
RAPTOR: Recursive Abstractive Processing for Tree-Organized Retrieval. RAPTOR introduces a novel approach to retrieval-augmented language models by constructing a recursive tree structure from documents.
2. What is the purpose of a raptor rag?
Unlike traditional RAG, RAPTOR organizes data in a tree structure, summarizing at each layer from the bottom up. This method captures broader context and enhances the representation of large-scale discourse, overcoming limitations of retrieving only short text chunks.
3. What is tree structured indexing and retrieval in Raptor?
A new and powerful indexing and retrieving technique for LLM in a comprehensive manner.
4. What is an advanced rag?
Advanced RAG helps LLM to avoid/reduce hallucinations. Advanced RAG enables embedding meta-data along with the documents and this helps LLMs with additional context resulting in improved generation. embedding Meta-Data is KEY for Advanced RAG.
5. What is RAPTOR in AI?
RAPTOR RAG is a method in AI for efficient, context-aware document retrieval using a recursive tree structure, enhancing retrieval-augmented models.
6. How to use rag with openai?
To use RAG with OpenAI, integrate OpenAI's API for language generation with a RAG model, fetching relevant documents from a knowledge base to augment responses for enhanced context and accuracy.
7. What is rag LangChain?
RAG (Retrieval-Augmented Generation) LangChain is a framework combining RAG with LangChain's capabilities to create advanced AI systems. It leverages document retrieval to enhance language models, improving context and accuracy in responses.
8. Does OpenAI have rag?
OpenAI does not have a native RAG (Retrieval-Augmented Generation) implementation. However, you can create a RAG system by integrating OpenAI's language models with external retrieval mechanisms, such as Elasticsearch or other document retrieval systems, to provide context-aware responses.
9. How to read an unstructured PDF in Python?
Firstly, we import the fitz module of the PyMuPDF library and pandas library. Then the object of the PDF file is created and stored in doc and the 1st page of the PDF is stored on page1. Using the PyMuPDF library to extract data from PDF with Python, the page. get_text() method extracts all the words from page 1.
10. What is an example of unstructured data?
Multimedia content: Digital photos, audio, and video files are all unstructured. Complicating matters, multimedia can come in multiple format files, produced through various means. For instance, a photo can be TIFF, JPEG, GIF, PNG, or RAW, each with their own characteristics.


